The results of the ABIDE Preprocessed initiative are currently available on a public Amazon S3 bucket. The data on S3 are stored as a single file per derivative for each participant, pipeline and strategy, which provides flexibility about the files that are downloaded. In the future, we hope to offer the data on NITRC as well with a tar file for each derivative, pipeline and strategy.
Accessing data from the Amazon S3 bucket
Each file in the S3 bucket can only be accessed using HTTP (i.e.,no ftp or scp). You must contruct a URL for each desired file (see URL templates below) and then download it using an HTTP client such as a web browser, wget, or curl. Each file can only be accessed using its literal name- wildcards will not work. An example python script for downloading a subset of the data based on participant demographics is available here (right click and select Save Link As...). You can find instructions for using this script here.
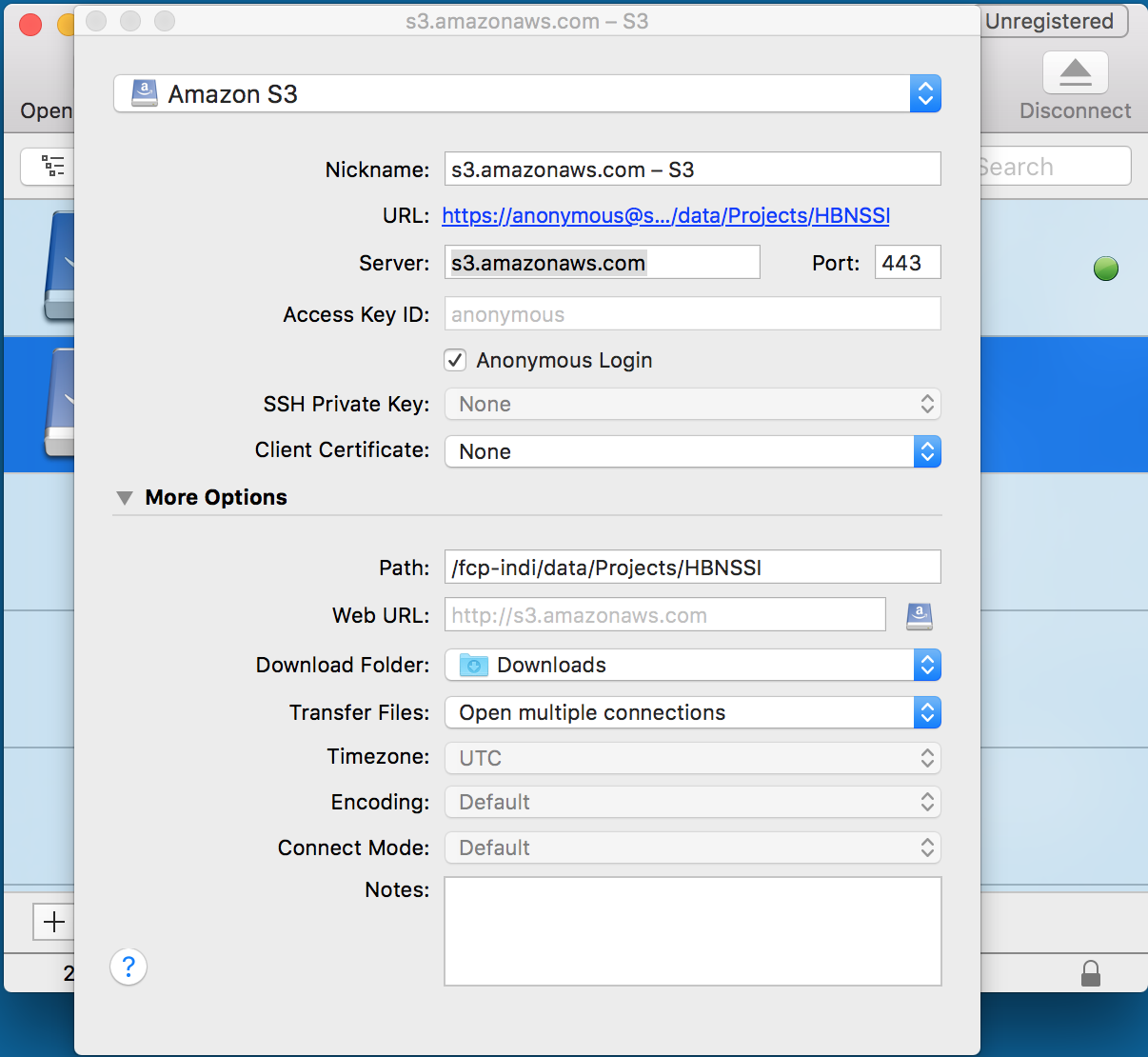
There are also file transfer programs that can handle S3 natively and will allow you to navigate through the data using a file browser. Cyberduck is one such program that works with Windows and Mac OS X (see screenshot illustrating a configuration to connect to the ABIDE preprocessed data below). Cyberduck also has a command line version that works with Windows, Mac OS X, and Linux.
Summary Spreadsheet
A summary spreadsheet that contains phenotypic data and quality assessment information is available here. This file contains all of the information from the original phenotypic file provided with the ABIDE release (which is described in the ABIDE phenotypic data legend), with additional metadata about the preprocessed data. This additional data includes:
- The
FILE_IDcolumn, which provides a mapping between the phenotypic data and the preprocessed data filenames. It is formed by combiningSITE_IDandSUB_ID(the latter of which is zero-padded to have 7 digits). It is used in every URL template described below. - Several columns that contain quality measures from the PCP Quality Assessment Protocol (
anat_cnr,anat_efc,anat_fber,anat_fwhm,anat_qi1,anat_snr,func_efc,func_fber,func_fwhm,func_dvars,func_outlier,func_quality,func_mean_fd,func_num_fd,func_perc_fd,func_gsr). For more detail click here. - Several columns that contain a description of the manual quality assessment annotations (
qc_rater_1,qc_notes_rater_1,qc_anat_rater_2,qc_anat_notes_rater_2,qc_func_rater_2,qc_func_notes_rater_2,qc_anat_rater_3,qc_anat_notes_rater_3,qc_func_rater_3, andqc_func_notes_rater_3). For more detail click here. - The
SUB_IN_SMPcolumn indicates whether the data was included in (Di Martino et al. 2014).
Functional Data URL Template
Preprocessed functional data can be downloaded using the following template:
https://s3.amazonaws.com/fcp-indi/data/Projects/ABIDE_Initiative/Outputs/[pipeline]/[strategy]/[derivative]/[file identifier]_[derivative].[ext]
Where:
[pipeline] = ccs | cpac | dparsf | niak
[strategy] = filt_global | filt_noglobal | nofilt_global | nofilt_noglobal
[file identifier] = the FILE_ID value from the summary spreadsheet
[derivative] = alff | degree_binarize | degree_weighted | dual_regression | ...
eigenvector_binarize | eigenvector_weighted | falff | func_mask | ...
func_mean | func_preproc | lfcd | reho | rois_aal | rois_cc200 | ...
rois_cc400 | rois_dosenbach160 | rois_ez | rois_ho | rois_tt | vmhc
[ext] = 1D | nii.gz
The file extension is determined by the derivative type. Use .nii.gz for all derivatives except for the ROI time series files, which end in .1D (these derivative names begin with rois_). Refer to the ROI description for more information about the definition of the ROIs used to extract these time series and their labels.
Here are a few examples that illustrate the construction of paths for a few different files:
ALFF for OHSU_0050147 preprocessed using filt_global from C-PAC (link):
https://s3.amazonaws.com/fcp-indi/data/Projects/ABIDE_Initiative/Outputs/cpac/filt_global/alff/OHSU_0050147_alff.nii.gz
Harvard-Oxford ROI time series for KKI_0050822 preprocessed using filt_global from C-PAC (link):
https://s3.amazonaws.com/fcp-indi/data/Projects/ABIDE_Initiative/Outputs/cpac/filt_global/rois_ho/KKI_0050822_rois_ho.1D
The 3D binary derivatives (i.e. those ending in nii.gz except for ‘func_preproc’ and ‘dual_reg’) are roughly 256 KB to 512KB in size. The ‘dual_reg’ files are 10 times the size of the others (i.e. 2.5MB - 5MB) and the ‘func_preproc’ files are very large (30 MB - 200 MB). Extracted time series files are .5 - 1 MB in size.
Minimally Preprocessed Data URL Template
Minimally preprocessed data using the C-PAC pipeline is available. These data can be downloaded from using the following template:
https://s3.amazonaws.com/fcp-indi/data/Projects/ABIDE_Initiative/Outputs/cpac/func_minimal/[file identifier]_func_minimal.nii.gz
Where:
[file identifier] = the FILE_ID value from the summary spreadsheet
For example, the URL for minimally preprocessed data for OHSU_0050147 (link) would be:
https://s3.amazonaws.com/fcp-indi/data/Projects/ABIDE_Initiative/Outputs/cpac/func_minimal/OHSU_0050147_func_minimal.nii.gz
Structural Data URL Templates
Due to the diversity of the structural pipelines, each pipeline has a different format for specifying its derivatives. The aforementioned Python script also provides examples for downloading this data. Again, file identifiers correspond to FILE_ID values from the summary spreadsheet.
ANTS Cortical Thickness URL Templates
Cortical thickness measures calculated using the ANTs pipeline can be downloaded using the root URL:
https://s3.amazonaws.com/fcp-indi/data/Projects/ABIDE_Initiative/Outputs/ants/
appended with a string corresponding to one of the two available cortical thickness derivatives:
-
A 3D volume containing voxel-wise measures of cortical thickness:
anat_thickness/[file identifier]_anat_thickness.nii.gz -
A text file containing average cortical thickness values for cortical regions of interests (ROIs):
roi_thickness/[file identifier]_roi_thickness.txt
ROIs were defined using sulcus landmarks according to the Desikan-Killiany-Tourville (DKT) protocol1 using the OASIS-TRT-20 joint fusion atlas in OASIS-30 space. Labels corresponding to these ROIs can be found here.
CIVET URL Templates
More information about CIVET can be found at its documentation page, which includes a description of output files.
Surfaces
CIVET generated surfaces in stereotaxic space can be downloaded using the following template:
https://s3.amazonaws.com/fcp-indi/data/Projects/ABIDE_Initiative/Outputs/civet/surfaces_[surface]/[file identifier]_[surface].obj
Where:
[file identifier] = the FILE_ID value from the summary spreadsheet
[surface] = gray_surface_rsl_left_81920 | gray_surface_rsl_right_81920 | mid_surface_rsl_left_81920 | ...
mid_surface_rsl_right_81920 | white_surface_rsl_left_81920 | white_surface_rsl_right_81920
Vertex-based Measures
Vertex-based measures in stereotaxic space can be downloaded using the following template:
https://s3.amazonaws.com/fcp-indi/data/Projects/ABIDE_Initiative/Outputs/civet/surfaces_[derivative]/[file identifier]_[derivative].txt
Where:
[file identifier] = the FILE_ID value from the summary spreadsheet
[derivative] = mid_surface_rsl_left_native_area_40mm | mid_surface_rsl_right_native_area_40mm | ...
native_pos_rsl_asym_hemi | surface_rsl_left_native_volume_40mm | surface_rsl_right_native_volume_40mm
Region-based Measures
Region-based measures can be downloaded using the following template:
https://s3.amazonaws.com/fcp-indi/data/Projects/ABIDE_Initiative/Outputs/civet/surfaces_[derivative]/[file identifier]_[derivative].dat
Where:
[file identifier] = the FILE_ID value from the summary spreadsheet
[derivative] = gi_left | gi_right | lobe_areas_40mm_left | lobe_areas_40mm_right | lobe_native_cortex_area_left | ...
lobe_native_cortex_area_right | lobe_thickness_tlink_30mm_left | lobe_thickness_tlink_30mm_right | ...
lobe_volumes_40mm_left | lobe_volumes_40mm_right
Cortical Thickness Maps
Cortical thickness maps in stereotaxic space can be downloaded using the following template:
https://s3.amazonaws.com/fcp-indi/data/Projects/ABIDE_Initiative/Outputs/civet/thickness_[derivative]/[file identifier]_[derivative].txt
Where:
[file identifier] = the FILE_ID value from the summary spreadsheet
[derivative] = cerebral_volume | native_rms_rsl_tlink_30mm_asym_hemi | native_rms_rsl_tlink_30mm_left | ...
native_rms_rsl_tlink_30mm_right
Freesurfer URL Template
The entirety of the Freesurfer output folders for each subject are available for download using the following template:
https://s3.amazonaws.com/fcp-indi/data/Projects/ABIDE_Initiative/Outputs/freesurfer/5.1/[file identifier]/[sub directory]/[output file]
Where:
[file identifier] = the FILE_ID value from the summary spreadsheet
[sub directory] = one of the standard Freesurfer subdirectories: label | mri | scripts | stats | surf
[output file] = the name of the desired output file
There are 284 files distributed across the subdirectories of each subject’s output directory. An example listing of the files for one subject is available here (right click and select Save Link As ...). The -qcache flag was used during reconstruction resulting in versions of the different surface metrics that have been smoothed at 0, 5, 10, 15, 20 and 25 mm FWHM. Information about the different subdirectories and files can be found in the Freesurfer documentation:
- ReconAllOutputFiles
- ReconAllTableStableV5.1
- ReconAllFilesVsSteps
- Anatomical ROI analysis
- Inspection of Freesurfer Output
- FreeSurfer File Formats
References
-
Klein, A. and Tourville, J., 2012. 101 labeled brain images and a consistent human cortical labeling protocol. Frontiers in Brain Imaging Methods. 6:171. DOI: 10.3389/fnins.2012.00171. ↩